GLMol A-Frame port
Old small test (1MBN) with A-Frame 0.8.2,
new test with A-Frame 1.0.4 2POR and
cid 31703.
No Oculus Go controls! tested 6 June
Use touchpad or joystick for navigation. You can "fly" around a model.
It is necessary to rotate your head too (but it is useful for health :)
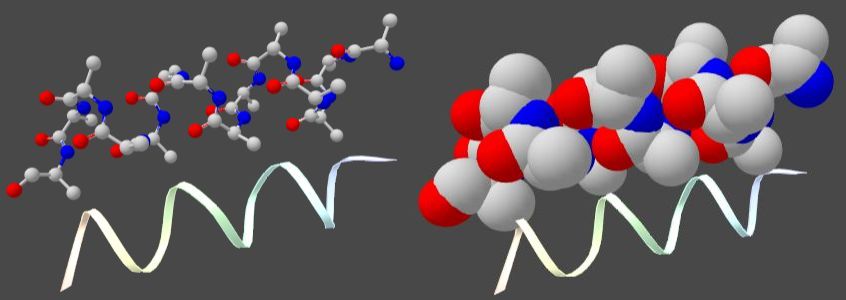
Protein α-helix structure is made by H-bonds
between nitrogen and oxygen atoms at the two nearest turns of the spiral
(between blue and red balls in the figure to the right).
See also wikipedia.org/wiki/Peptide_bond and
en.wikipedia.org/wiki/Protein_secondary_structure.
You can see from α-helix in sphere mode,
that it is not a real spiral, but a tightly packed spiral structure.
Spiral bands are used just to show protein molecule structure.
PDB file thanks to V.Tsvetkov from www.bashedu.ru.
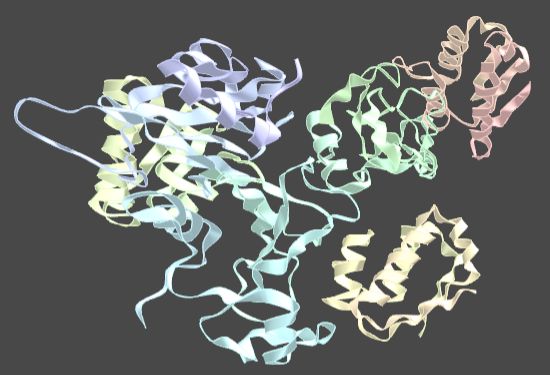
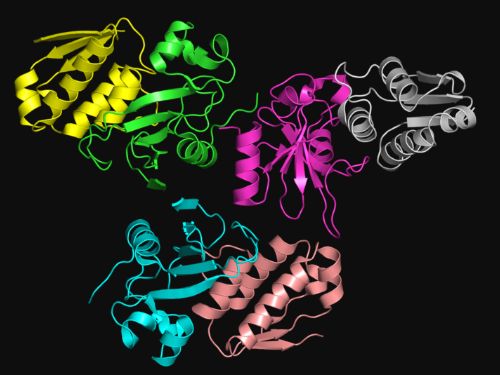
Protein interaction demo (see
Water-mediate interaction at aprotein-protein interface).
For some reason the model includes 3 pairs of molecules matching each other.
A few big molecules to test VR usefulness and performance:
PDB 6VAK, 6AUI
(is too big and slow for Oculus Go).
RCSB PDB server: 6LU7, Paclitaxel 1JFF, 1H69, 4CR2
(can't plot VERY BIG molequles e.g. 6OM0).
PubChem CID: Doxorubicin 31703, chlorogenic acid 1794427, 2335.
Old big VR GLMol test does not work in VR for me but
You can load and see in VR any files: RCSB PDB or
PubChem CID.
I will try to add local files download on your request.
Comments
Navigation in small tests works thanks to Brandon and Meduzo from Discord #aframe.
I failed yet to make model rotation controls.
Model movement doesn't work in Oculus browser with A-Frame > 1.0.0
(I made an issue,
1.0.0 box test).
For quick tests one can use
repl.it/@meduzo/molview.
Camera look-controls works in A-Frame 1.0.4 with VR BOX gamepad. So is it Oculus feature?
The application is modifed a little to load files from URL.
We need an expert to make interesting models with animations and explanations!
Very simple animation demo.
Animations
GLmol A-frame port now can generate large biomolecule VR models.
There are multi-frame PDB data files. So in principle, it is possible
to make animations by model interpolation between frames.
To avoid data transfer every frame one could morph meshes in a shader.
Animations
made with UCSF Chimera:
"Folding of the engrailed homeodomain" One 1ENH frame has only 500 atoms (~40kb).
For a keyframe animation (Aframe + ThreeJS). Can I morph model between
two samples (with similar meshes) in a shader on GPU, i.e. aPos = aPos1*t + aPos2*(1-t)?
I've found THREEjs "morphing shaders" at
https://threejsfundamentals.org/threejs/lessons/threejs-optimize-lots-of-objects-animated.html
how to use that in A-frame next :)
Links
github.com/SBUtltmedia/aframe-glmol-component
Protein Database Viewer it.stonybrook.edu
My old (2012) WebGL demo
Design and Architecture of GLmol
nanome.ai
Carbon

Fullerene ("sphere" mode).
Carbon nanotube ("sphere" mode).
4 tubes are too heavy for Oculus Go.
Diamond lattice.
Graphite lattice or may be 4 graphene layers :)
WebXR samples
updated 6 June 2020








